Description
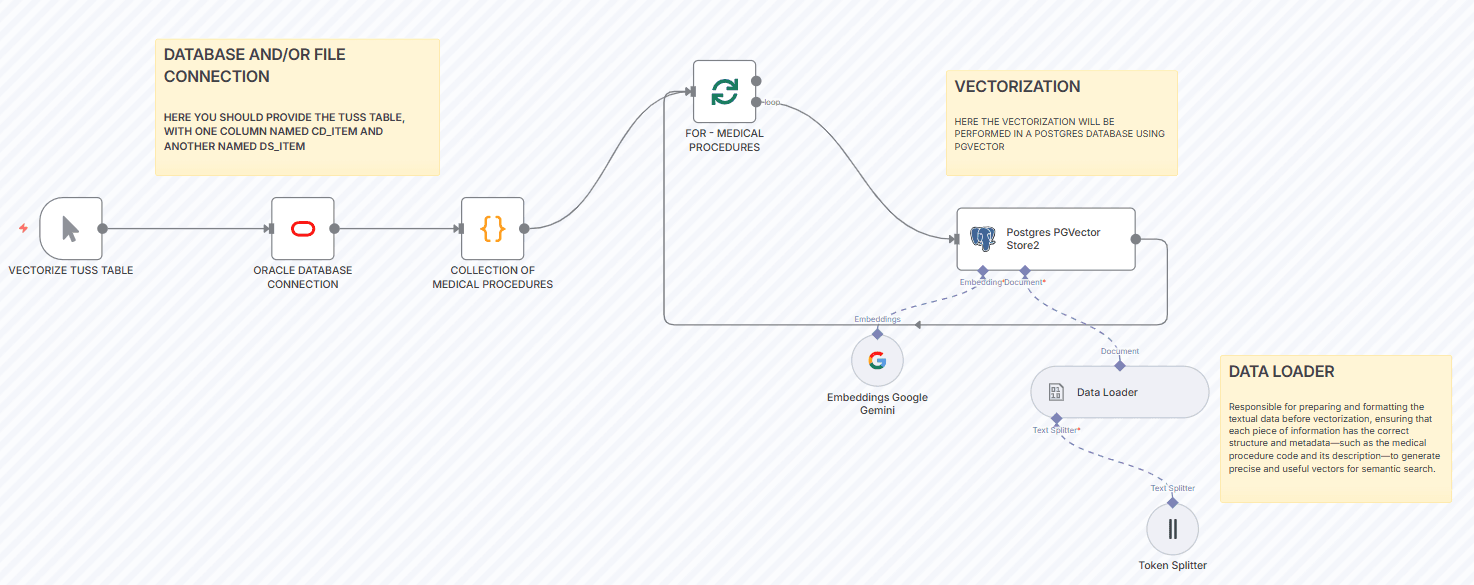
This workflow vectorizes the TUSS (Terminologia Unificada da Saúde Suplementar) table by transforming medical procedures into vector embeddings ready for semantic search.
It automates the import of TUSS data, performs text preprocessing, and uses Google Gemini to generate vector embeddings. The resulting vectors can be stored in a vector database, such as PostgreSQL with pgvector, enabling efficient semantic queries across healthcare data.
What Problem Does This Solve?
Searching for medical procedures using traditional keyword matching is often imprecise. This workflow enhances the search experience by enabling semantic similarity search, which can retrieve more relevant results based on the meaning of the query instead of exact word matches.
How It Works
- Import TUSS data: Load medical procedure entries from the TUSS table.
- Preprocess text: Clean and prepare the text for embedding.
- Generate embeddings: Use Google Gemini to convert each procedure into a semantic vector.
- Store vectors: Save the output in a PostgreSQL database with the pgvector extension.
Prerequisites
- An n8n instance (self-hosted).
- A PostgreSQL database with the pgvector extension enabled.
- Access to the Google Gemini API.
- TUSS data in a structured format (CSV, database, or API source).
Customization Tips
You can adapt the preprocessing logic to your own language or domain-specific terms.
Swap Google Gemini with another embedding model, such as OpenAI or Cohere.
Adjust the chunking logic to control the granularity of semantic representation.
Setup Instructions
Prepare a source (database or CSV) with TUSS data. You need at least two fields:
-
CD_ITEM (Medical procedure code)
-
DS_ITEM (Medical procedure description)
Configure your Oracle or PostgreSQL database credentials in the Credentials section of n8n.
Make sure your PostgreSQL database has pgVector installed.
Replace the placeholder table and column names with your actual TUSS table.
Connect your Google Gemini credentials (via OpenAI proxy or official connector).
Run the workflow to vectorize all medical procedure descriptions.